Understanding the contribution of different genes to the fitness of individual patient tumours remains a considerable research and clinical challenge. A gene driving growth in one tumour subtype may be neutral or even protective in another. Even within the same subtype, the dependency of a tumour on a gene mutation may be influenced by interactions with mutations in many other genes. Furthermore, these dependencies change as the tumour evolves in time, during spread to different cellular environments, and in response to therapy. Large-scale tumour sequencing programmes have contributed greatly to improved understanding of gene function in different tumour backgrounds. Nonetheless, the sheer complexity delivered by tens of thousands of mutations in an individual patient tumour still limits our ability to determine the fitness of genes, and consequently to predict with confidence the outcome of alternate therapy choices.
As reported in Nature Communications1, a team at BC Cancer and UBC has developed a methodology to accurately quantify the fitness of panels of genes in human tumours propagated in mice as patient-derived xenografts (PDXs). Led by Principal Investigators Sam Aparicio, Alexandre Bouchard Côté and Andrew Roth and lead author Peter Eirew, the team focused on a set of 21 patient breast tumour PDX lines, including 13 from triple negative subtypes for which targeted treatment options are often lacking.

Pooled CRISPR screens have previously been carried out in cancer cell lines in culture, but not in PDXs growing in vivo. The in vivo application brings powerful benefits, notably the ability to assess gene fitness in a physiological setting, and in a wide range of patient-specific tumour genotypes. However, there are particular challenges posed by the biology of PDX growth, in which the contribution of different transplanted cell clones to the final tumour mass is highly variable. To address this, the methodology incorporates lineage-tracking barcodes into the CRISPR vector. The statistical model then takes into account measured distributions of clone sizes within each regenerated tumour when inferring gene fitness values.
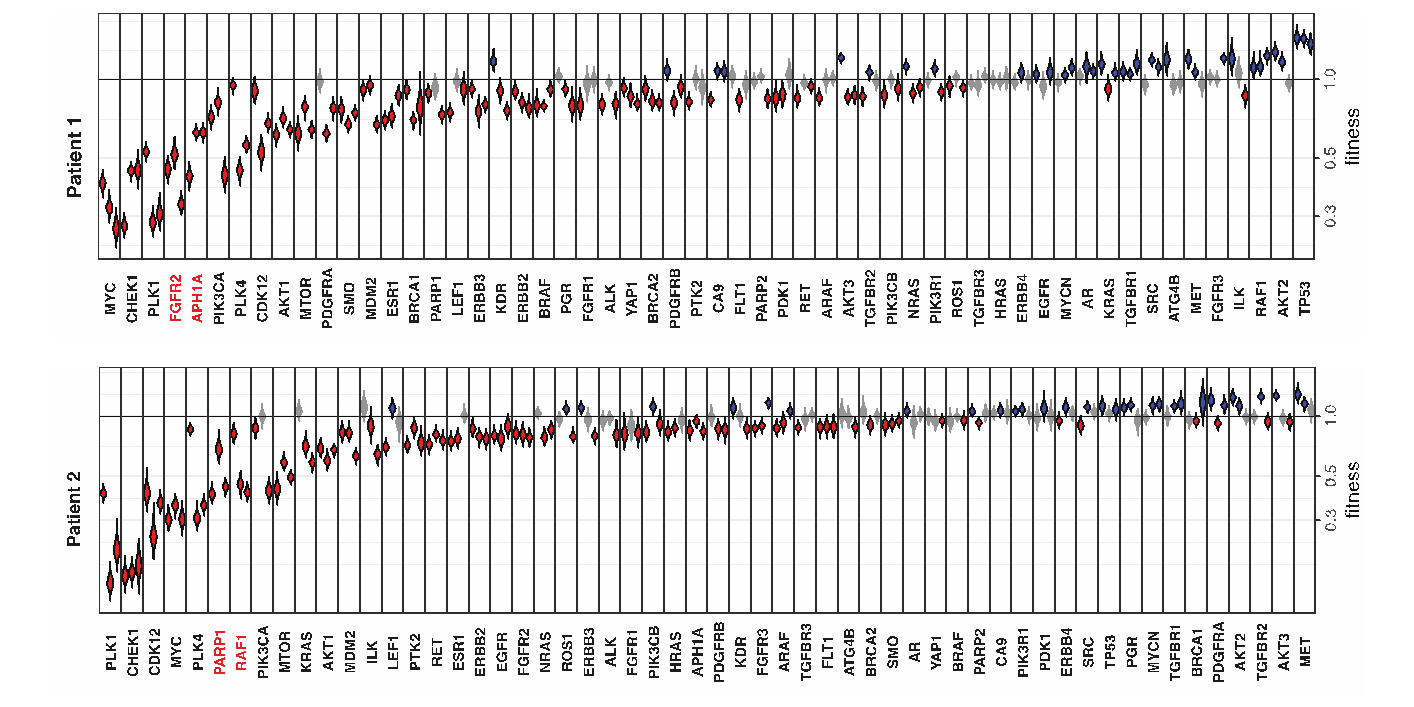
The study demonstrates a variability in the breast cancer fitness landscape dominated by biological differences between patient tumours, rather than by differences in the xenotransplantation site or whether the PDX line is used at an early or late passage. As an illustration, the figure shows the fitness profile of two different patient triple negative breast cancers assayed with a panel of breast cancer signaling genes, highlighting that the two tumours are vulnerable to disruption of different genes, including genes which are potential targets for therapy. The team also showed that the methodology can be used to probe in vivo activity of gene translocations, demonstrating the dependency of one patient tumour on a previously uncharacterized NOTCH3-MEMO1P4 fusion. Finally, combining the CRISPR fitness approach with the tractability of PDX models to drug studies, the team demonstrated the potential to screen for conditional drug-gene interactions in patient tumour-specific backgrounds. In a proof of principle, treatment of an MDM2-amplified PDX with an MDM2-inhibitor caused a shift in CRISPR-mediated fitness of the TP53 gene from neutral to positive, consistent with the known epistatic relationship between the two genes.
The study is a collaboration between the UBC Departments of Pathology and Laboratory Medicine and the Department of Statistics, and supported by generous funding from Terry Fox Research Institute, Canadian Cancer Society, CIHR, Breast Cancer Research Foundation, Cancer Research UK and Canada Foundation for Innovation.
Eirew, P., O’Flanagan, C., Ting, J. et al. Accurate determination of CRISPR-mediated gene fitness in transplantable tumours. Nat Commun 13, 4534 (2022). https://doi.org/10.1038/s41467-022-31830-2